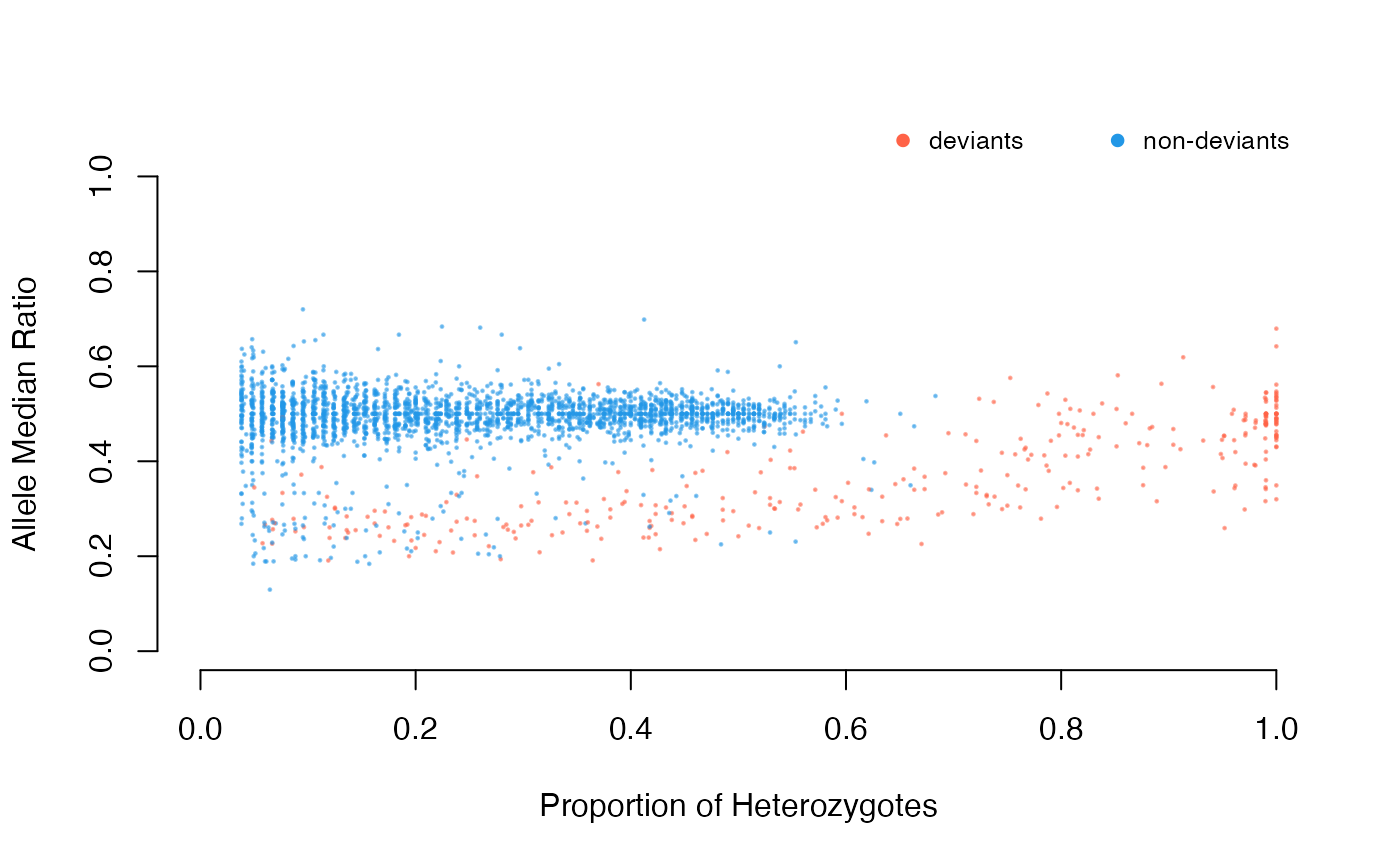
Detect deviant SNPs using excess of heterozygotes (alleles that do not follow HWE) and allelic-ratio deviations (alleles with ratios that do not follow a normal Z-score or chi-square distribution). See details.
Arguments
- data
data frame of the output of
allele.info- Fis
numeric. Inbreeding coefficient calculated using
h.zygosity()function- test
character. type of test to be used for significance. See details
- intersection
logical, whether to use the intersection of the methods specified in
test(if more than one)- method
character. method for testing excess of heterozygotes. Fisher exact test (
fisher) or Chi-square test (chi.sq)- plot
logical. whether to plot the detected singlets and duplicates on allele ratio vs. proportion of heterozygotes plot.
- verbose
logical. show progress
- ...
additional parameters passed on to
plot
Details
SNP deviants are detected with both excess of heterozygosity according to HWE and deviant SNPs where depth values fall outside of the normal distribution are detected using the following methods:
Z-score test \(Z_{x} = \sum_{i=1}^{n} Z_{i}\); \(Z_{i} = \frac{\left ( (N_{i}\times p)- N_{Ai} \right )}{\sqrt{N_{i}\times p(1-p)}}\)
chi-square test \(X_{x}^{2} = \sum_{i-1}^{n} X_{i}^{2}\); \(X_{i}^{2} = (\frac{(N_{i}\times p - N_{Ai})^2}{N_{i}\times p} + \frac{(N_{i}\times (1 - p)- (N_{i} - N_{Ai}))^2}{N_{i}\times (1-p)})\)
See references for more details on the methods
Users can pick among Z-score for heterozygotes (z.het, chi.het),
all allele combinations (z.all, chi.all) and the assumption of no
probe bias p=0.5 (z.05, chi.05)